Enhancing Collision Detection in Taichi for Particles with Diverse Sizes
This long-awaited post documents an experiemnt that I conducted two years ago. In the previous post, we explored a basic Discrete Element Method (DEM) model, utilizing Taichi's data structures to accelerate collision detection. However, this method had a limitation: it became inefficient with particles of widely varying sizes. This post aims to refine the algorithm by overcoming this limitation.
Recap: Accelerating collision detection with a fixed grid
Discrete element method (DEM) involves solving the time evolution of a massive number of particles. One algorithmic bottleneck is the detecting the collision of particles. If we have particles, a brute-force method need to check the possibility of collision between every pair of particles - an operation with time complexity . As collision can only happen if one particle is in the proximity of another, we can narrow down the search area by partitioning the computational domain into a fixed grid as shown below. Then we only need to examine the 3 x 3 cells with a given particle in the center cell. In this way, we can speed up the collision detection process.
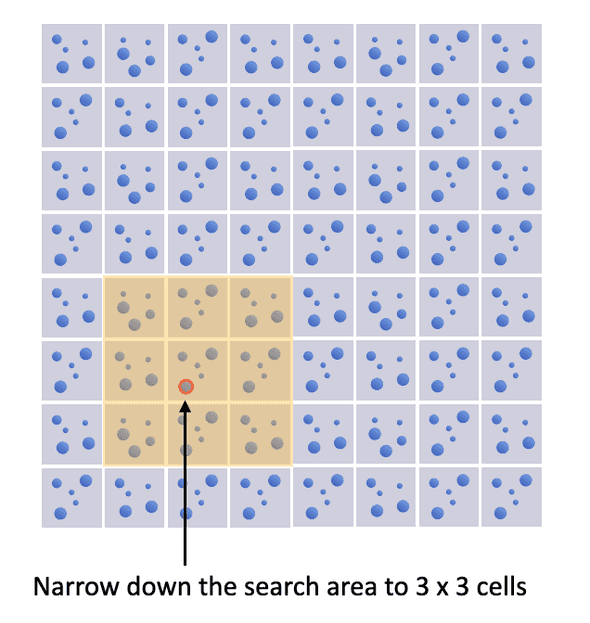
However, there's one caveat. We need to make sure the side length of a cell is greater than the diameter of the largest particle. Otherwise, a large particle in the center cell might collide with another large particle whose center is outside of the 3 x 3 cell region. As you can imagine, if we have a few extremely large particles in the domain with a massive number of small particles, the benefit of the grid would be limited. In particular, we have two straightforward options
- increase the side length of the grid cell to be greater than the diameter of the largest particle, so that we can still search within the 3 x 3 cells region.
- keep the grid cell small but expand the search region to be 5x5 or even larger.
Regardless of which approach we take, in the worse case scenario, the search region would be in the order of , resulting in a time complexity of for collision detection. This means the fixed grid has no advantage over brute force when the particle sizes are diverse. Fortunately, we can still leverage the grid structure with smaller grid cells by grouping particles by sizes and treating different groups of particles differently.
Enhanced Approach for Diverse Particle Sizes
This idea is described in the paper MR linear contact detection algorithm as as "multi-step collision detection". To illustrate the idea, we use an example as shown in the following example, where we have 4 large particles and 8192 small particles.

Simulation of 4 large particles and 8192 small particles
In this simple example, we group particles into two categories by size - small and large. Thus, there are three types of collision types possible - small-small, small-large and large-large. The key idea of the enhanced approach is to keep the grid cell small and vary the search region depending on the center particle:
- if the center particle is a small particle, we search in the 3 x 3 cell region
- if the center particle is a large particle, we search in a larger region, where the region should cover all possible collisions (the side length of the region should be greater than twice the diameter of the largest particle.)
One can easily extend this approach to multiple groups of particles, e.g. small, medium and large. Please refer to the MR linear contact detection algorithm paper for a detailed formulation.
This approach is referred to as "multi-step collision detection" in the paper since collisions are proposed to be resolved sequentially from the group of particles with the smallest sizes to the group with largest sizes. By implementing the algorithm in Taichi, we can parallelize these sequential steps. Thus, calling the approach as "multi-step collision detection" would be confusing in this post.
You can find the implementation using Taichi in https://github.com/DesmondZhong/dem/. The core block of code is shown below.
# Fast collision detectionfor i in range(n):if i < ns: # small particlesgrid_idx = ti.floor(gf[i].p * grid_n, int)# small particles only search neighboring 3x3 grid# to resolve small-small collisionx_begin = max(grid_idx[0] - 1, 0)x_end = min(grid_idx[0] + 2, grid_n)y_begin = max(grid_idx[1] - 1, 0)y_end = min(grid_idx[1] + 2, grid_n)for neigh_i in range(x_begin, x_end):for neigh_j in range(y_begin, y_end):neigh_linear_idx = neigh_i * grid_n + neigh_jfor p_idx in range(list_head[neigh_linear_idx],list_tail[neigh_linear_idx]):j = particle_id[p_idx]if j < ns and i < j:resolve(i, j) # small-small collisionelse: # large particlesgrid_idx = ti.floor(gf[i].p * grid_n, int)# large particles resolve all possible collisions# so they need to search in a larger regionx_begin = max(grid_idx[0] - SEARCH_NUM_GRID, 0)x_end = min(grid_idx[0] + 1 + SEARCH_NUM_GRID, grid_n)y_begin = max(grid_idx[1] - SEARCH_NUM_GRID, 0)y_end = min(grid_idx[1] + 1 + SEARCH_NUM_GRID, grid_n)for neigh_i in range(x_begin, x_end):for neigh_j in range(y_begin, y_end):neigh_linear_idx = neigh_i * grid_n + neigh_jfor p_idx in range(list_head[neigh_linear_idx],list_tail[neigh_linear_idx]):j = particle_id[p_idx]if j >= ns and i < j: # large-large collisionresolve(i, j)elif j < ns: # large-small collisionresolve(i, j)
Performance
To verify that the enhanced approach indeed boost performance empirically. We conduct experiemnts with different cell size and report the wall clock time below.
| method | ungrouped | grouped | grouped | grouped | grouped |
|---|---|---|---|---|---|
| grid_n | 5 | 5 | 32 | 64 | 128 |
| wall clock time (on gpu) | 37.41s | 40.48s | 35.21s | 33.66s | 37.06s |
Here, grid_n indicates the cell size. As we have a square domain, a grid_n of 5 means we divide the domain into 5 x 5 cells. We can see that if we choose the right grid_n (64 in this case), grouping the particles ("grouped") outperforms the baseline implementation ("ungrouped") where we keep the cell size large enough so that can still search the 3 x 3 region.
Conclusion
This post revisits my experiments on optimizing collision detection in DEM by categorizing particles by size. Inspired by the MR linear contact detection algorithm paper, I adapted the concept to a parallelized version using Taichi. Feel free to check out the codebase for more details.